RummaGEO Documentation
1.1 Gene Set Search
The Gene Set Search page enables users to search the RummaGEO database for gene sets that match their query gene set. Similarity to gene sets contained within the RummaGEO database with the query gene set is measured with Fisher's exact test. Any significantly overlapping gene sets are returned to the user along with their accompanying metadata. User query gene sets can be pasted or typed into the input form with each gene on a new line, or the user may upload a file containing genes where the genes are listed with new line, tab, or comma separators. Based on the gene symbols within the query gene set, this query will run against either the collection of automatically generated human gene sets or mouse gene sets:
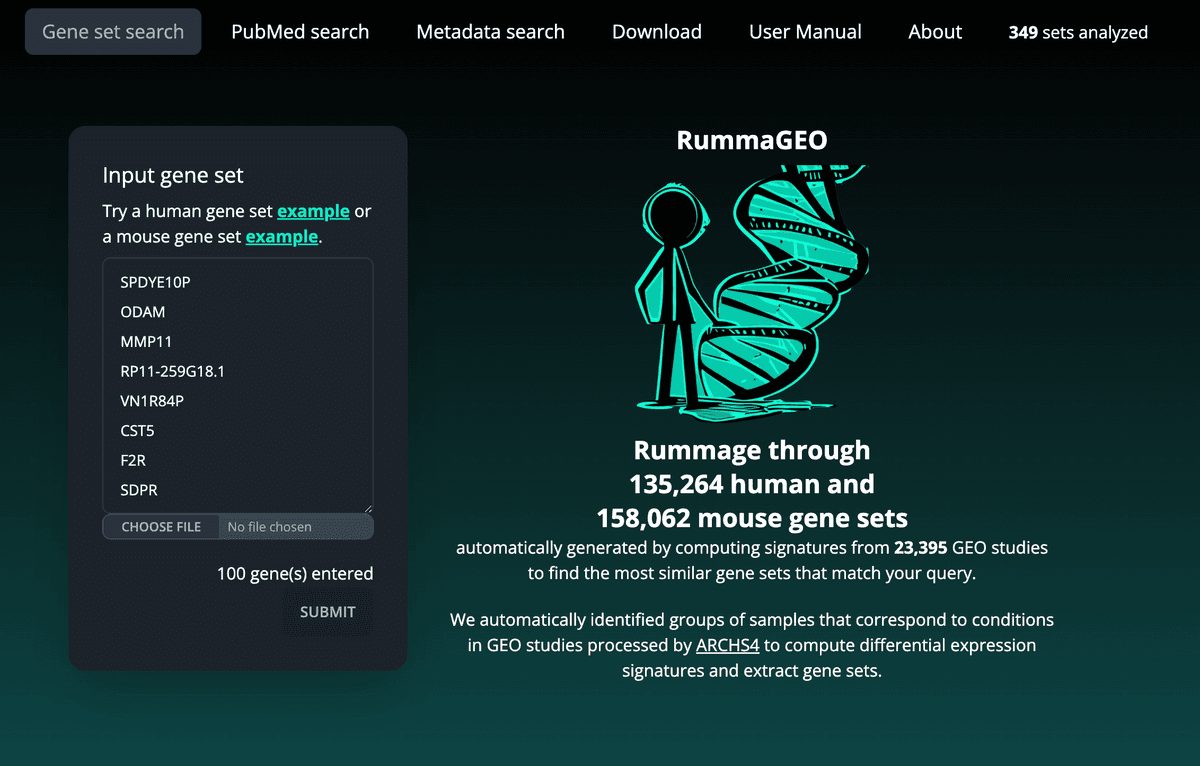
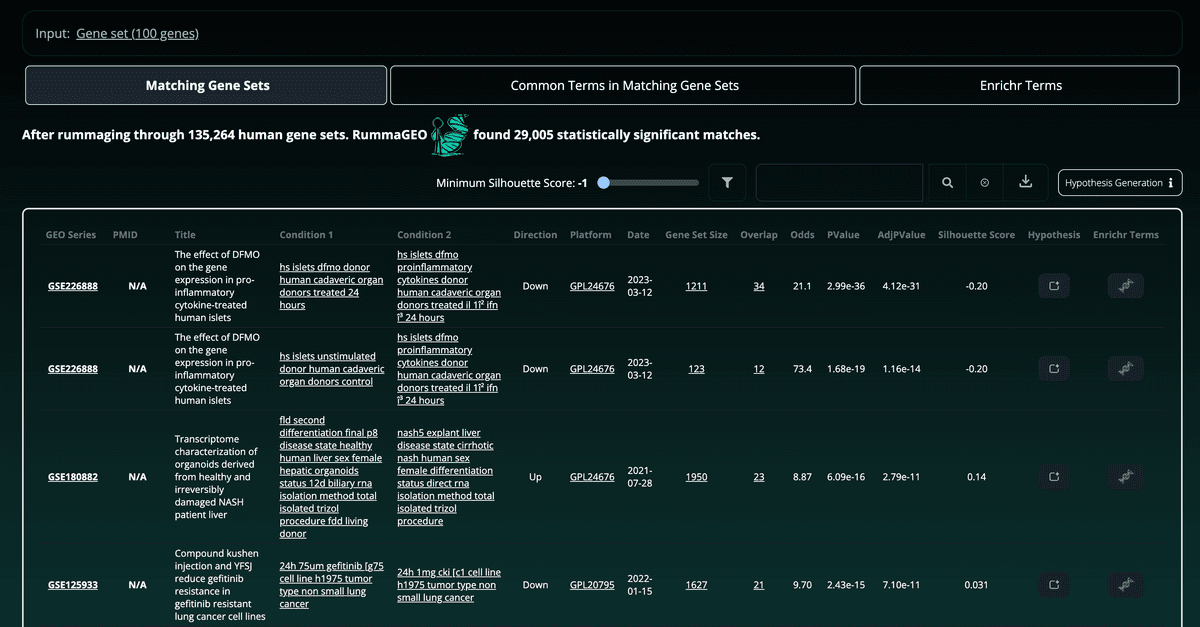
Paginated results are returned with the total number of gene sets which the query was compared to and the number of those gene sets which were significantly enriched. Enrichment statistics are provided on the right side of the table. The user may explore the metadata associated with the signatures on each results page, by inspecting the title of the GEO study, the corresponding linked GEO accession, and, if available, the linked PubMed ID:
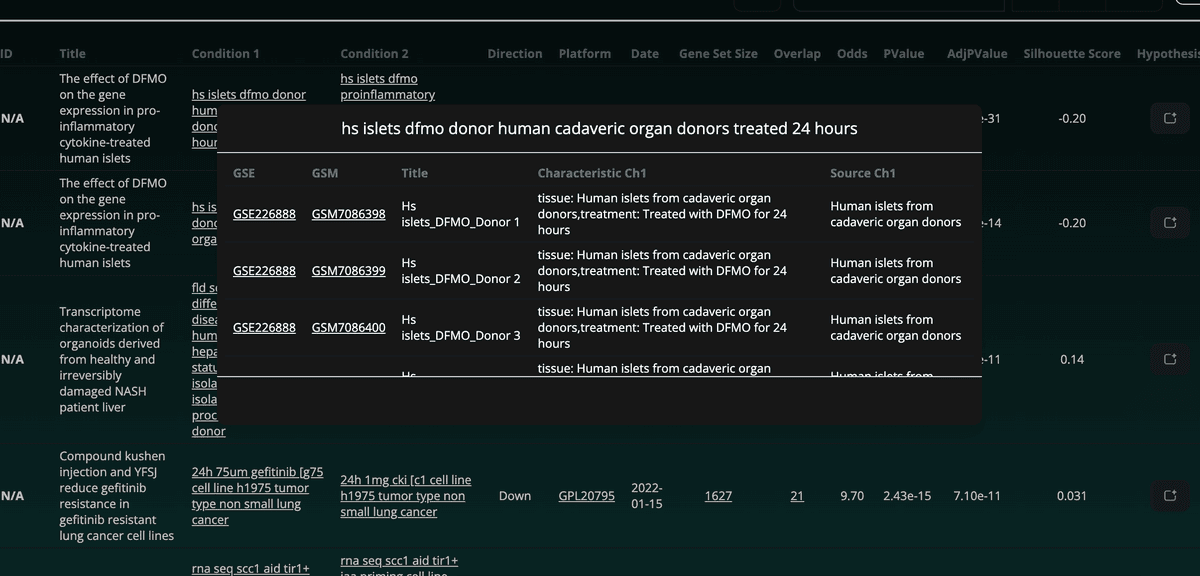
Additionally, the sample IDs (GSM) and their metadata associated with each condition are displayed in a modal box when clicked:
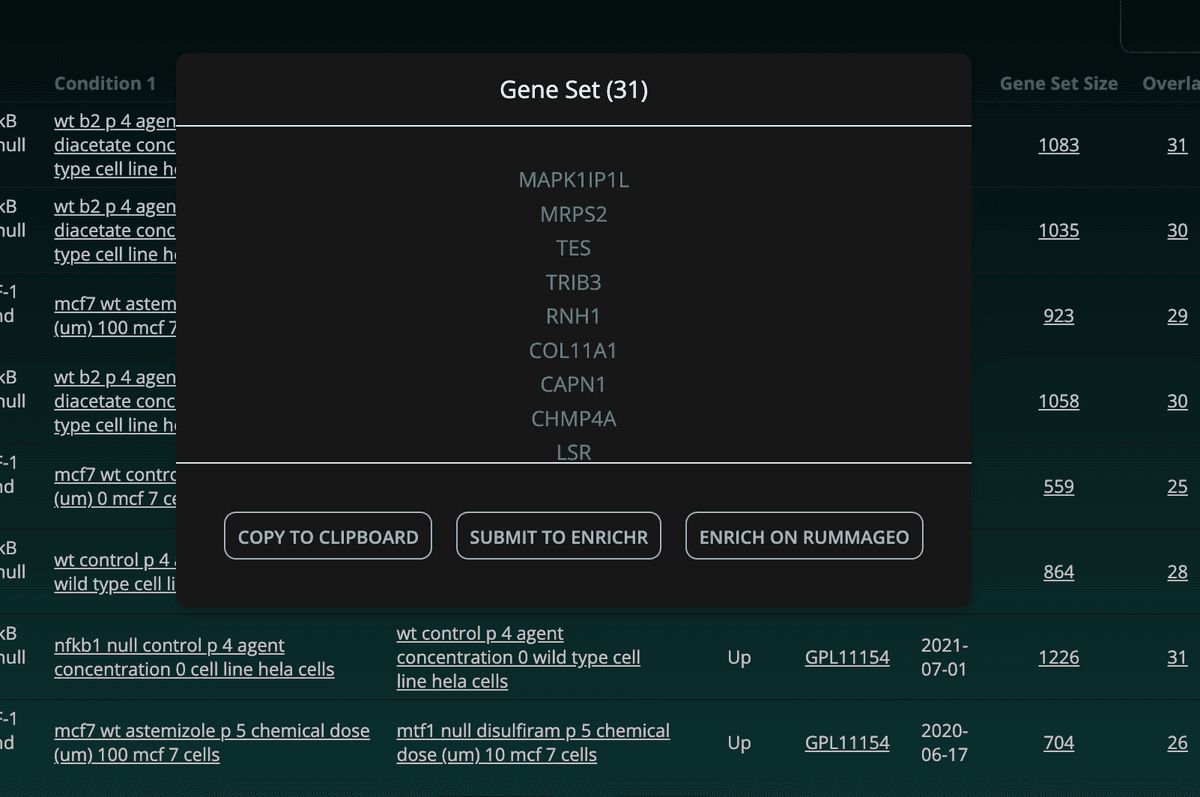
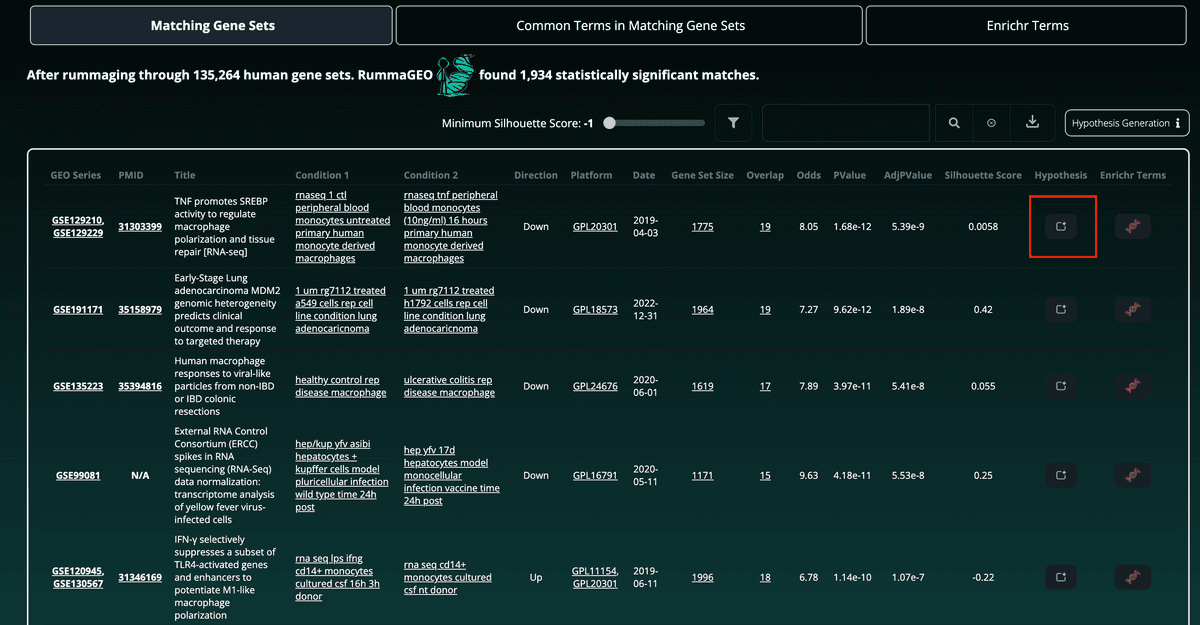
Other information such as the directionality of the gene set, the platform (GPL), the date of the study, the number of overlapping genes, and the total number of genes in the enriched gene set are also displayed. Clicking the overlap or gene set size will open a modal box with the corresponding genes as well as buttons to copy the gene set to the clipboard, or to view the enrichment results on RummaGEO or Enrichr:
To further filter and refine the results, the user may use the search bar located above the table to search for gene sets containing certain keywords. This allows, for instance, to view enriched results related to macrophages based upon the same input gene set. The total number of enriched gene sets is updated accordingly:
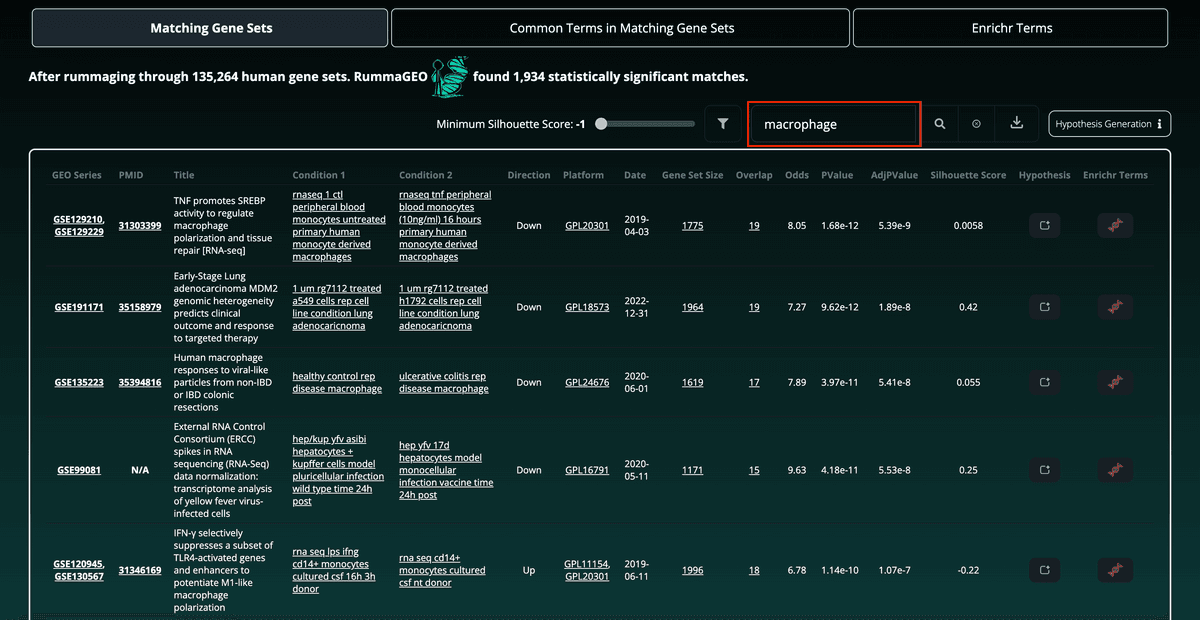
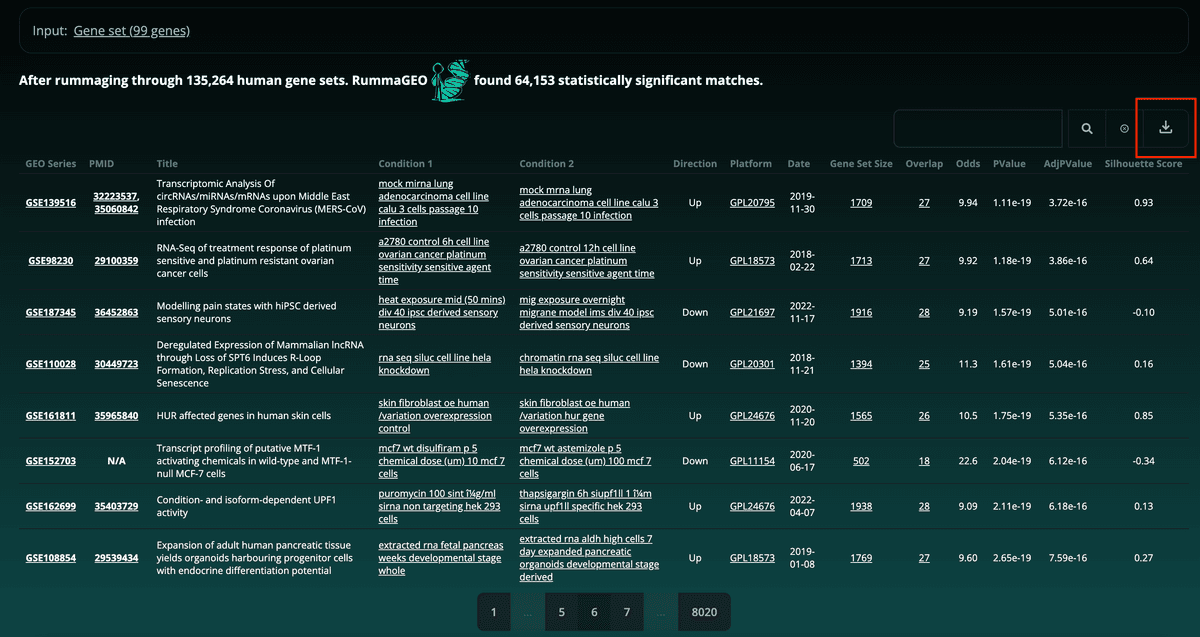
Results may also be easily downloaded using the button to the far right of the search feature in a tab delimited format:
Selecting the Common Terms in Matching Gene Sets tab will provide enrichment results computed with the Fisher's Exact Test for the first 5,000 unique GSEs from signatures returned in the gene set search result:
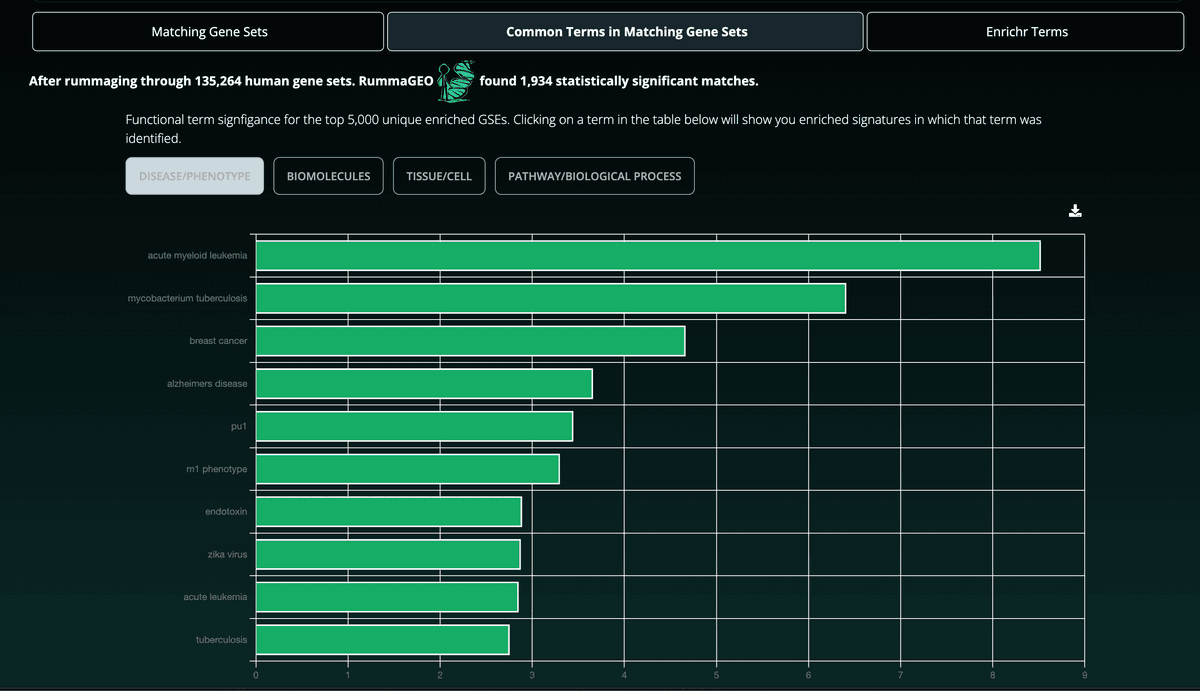
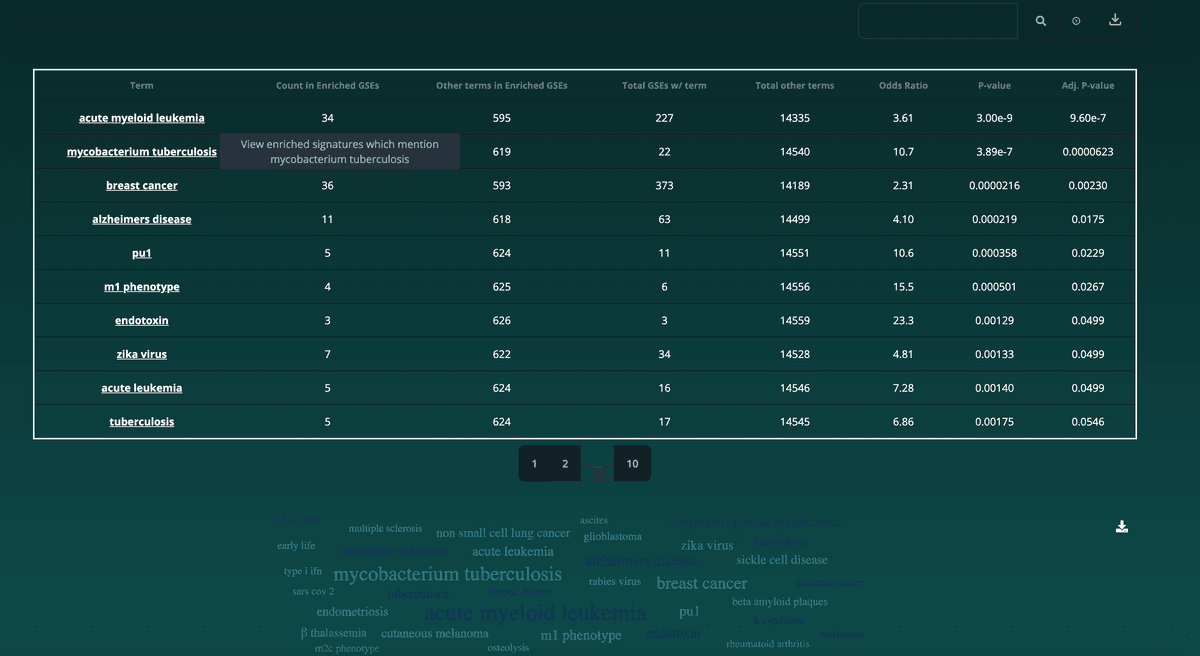
Additionally, below the bar chart, the user may view a table with all enriched terms including the signfigance of each term's appearance. Clicking a term will result in the user being brought back to the Matching Gene Sets tab wherein the user can view the gene sets that contain the selected term:
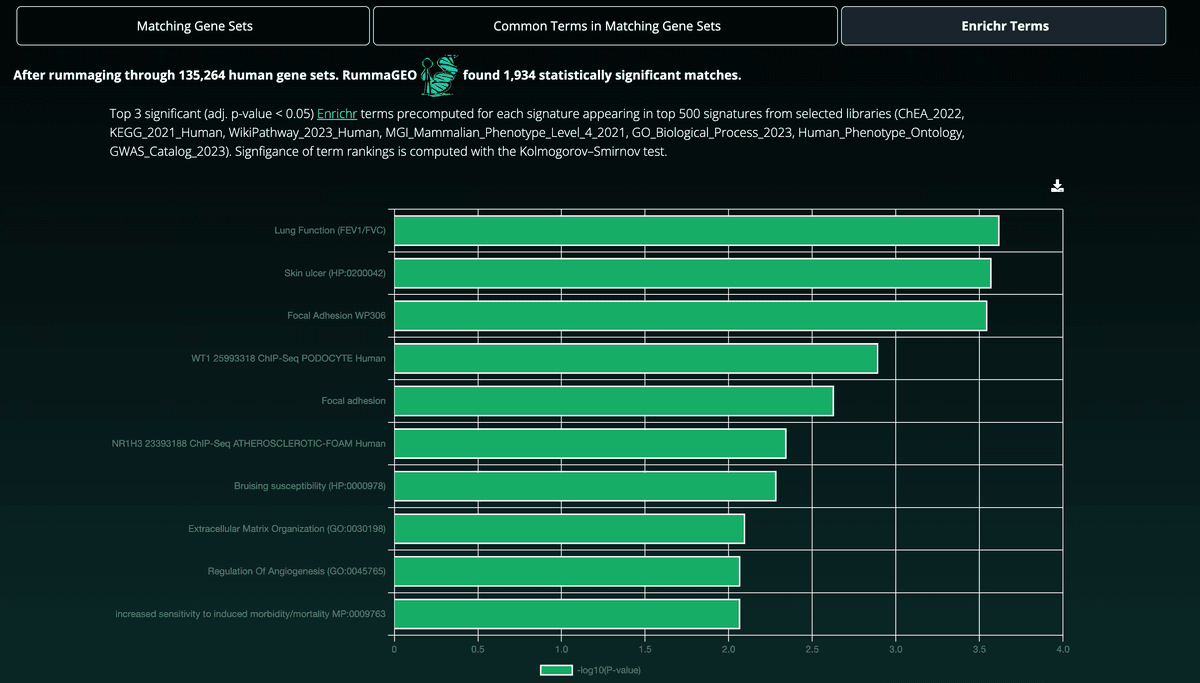
Selecting the Enrichr Terms tab will show the most commonly appearing Enrichr terms in the top 500 signatures returned from the gene set search. Enrichr terms are precomputed for all RummaGEO signatures for a selection of libraries (ChEA 2022, KEGG 2021 Human, WikiPathway 2023 Human, GO Biological Process 2023, MGI Mammalian Phenotype Level 4 2021, Human Phenotype Ontology, and GWAS Catalog 2023):
Hypothesis Generation
Users may also generate hypotheses concerning their gene set's overlap with any RummaGEO signatures:
After clicking the generate hypothesis button, users will need to enter a description of their gene set, which will be provided to the LLM:
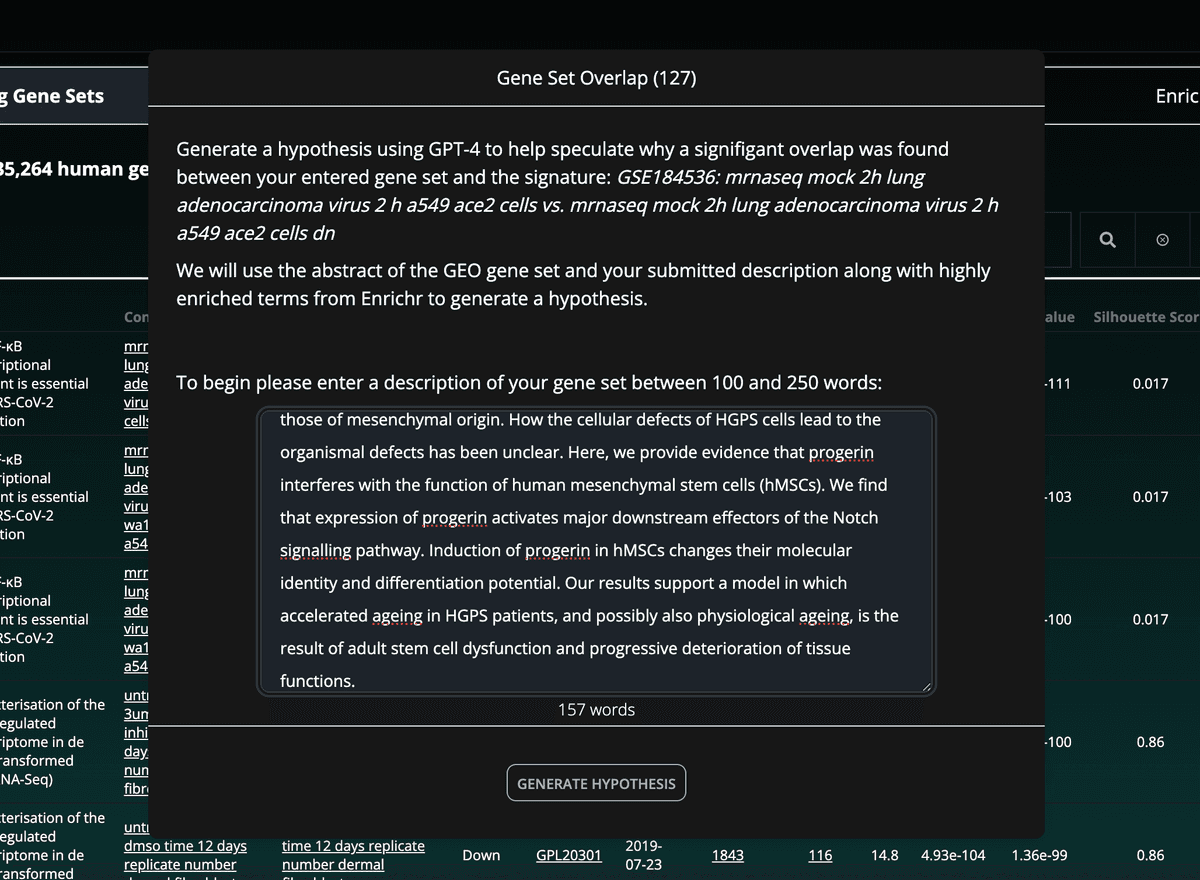
RummaGEO takes this description together with the matching RummaGEO gene set study abstract, and the top three significantly enriched terms from the overlapping genes from several Enrichr libraries (WikiPathway 2023 Human, GWAS Catalog 2023, GO Biological Process 2023, MGI Mammalian Phenotype Level 4 2021). The prompt additionally instructs the large language model (LLM) to reference all the provided descriptions and context of the gene sets, as well as the highly enriched terms from Enrichr. Hypotheses are then parsed to find references to any enriched terms, and insert the enrichment statistics as part of the hypothesis description:

1.2 PubMed Search
The PubMed Search page enables users to search for gene sets in RummaGEO based on a PubMed search using the PubMed API query. The top 5000 publications returned from the user's query are used to display extracted gene sets from the GEO studies associated with the returned papers. The number of articles returned by the PubMed API along with the number of associated gene sets and associated publications in the RummaGEO database are displayed at the top of the result:
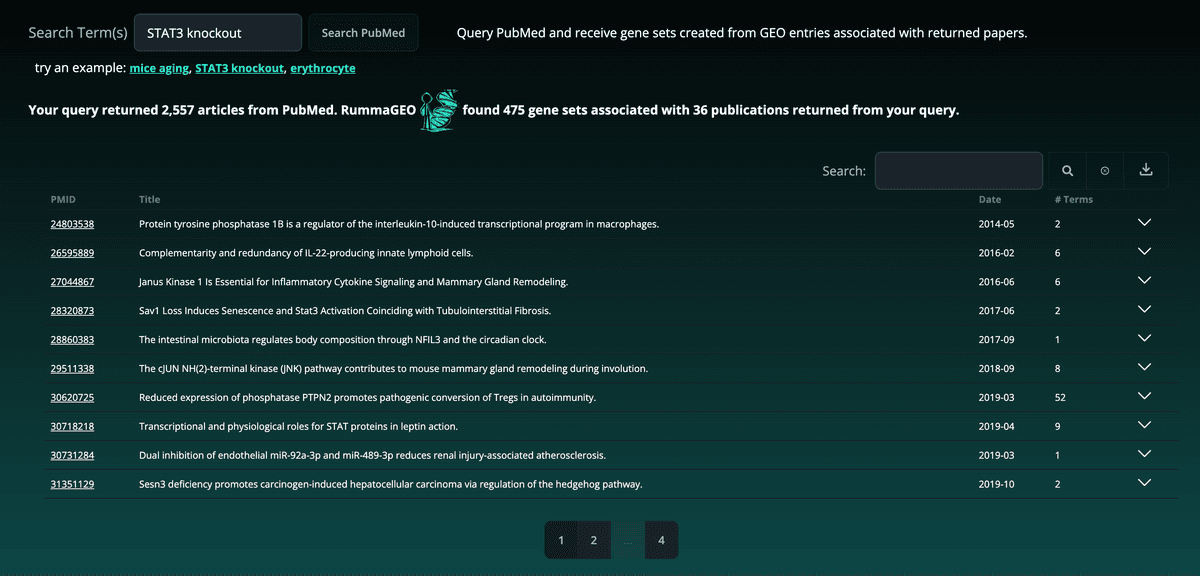
Paginated results are grouped by GEO study (GSE) with the corresponding signatures and available metadata located in a dropdown table:
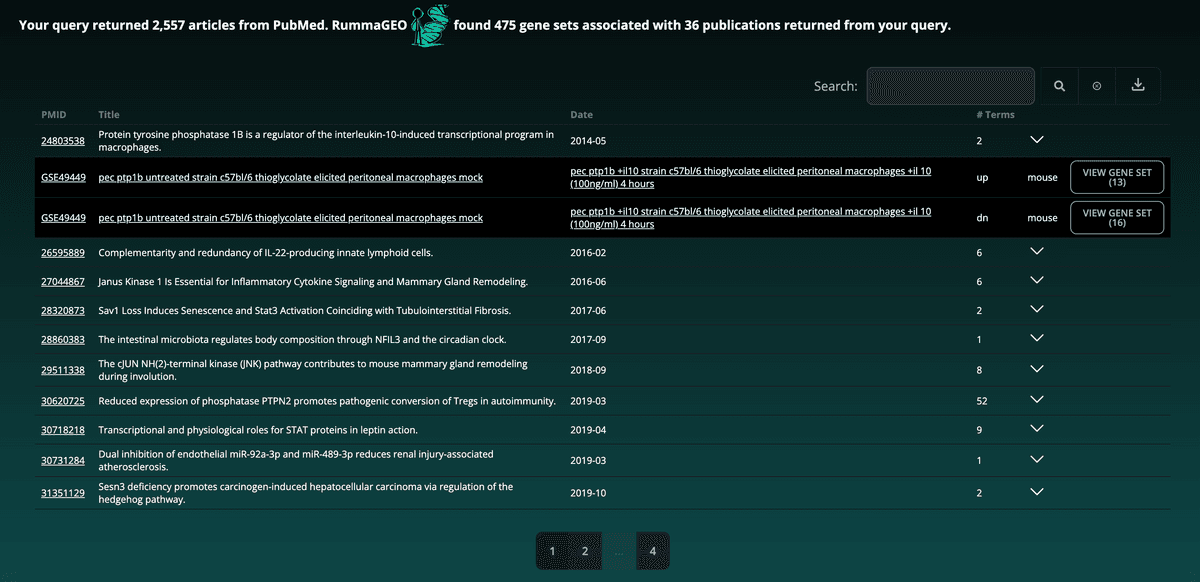
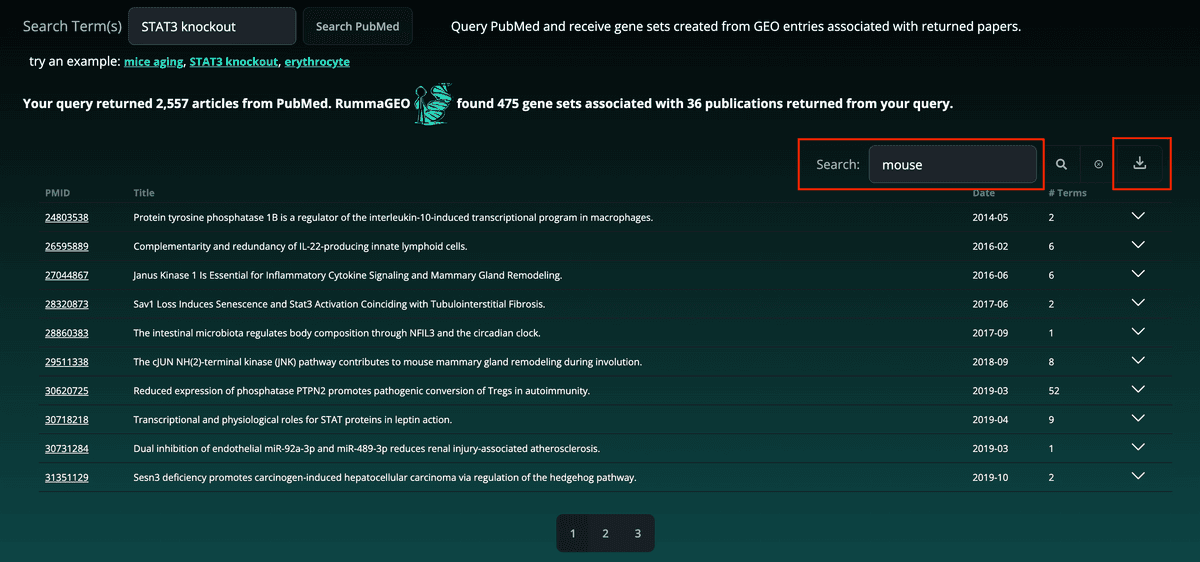
Additionally, the results can be further filtered using the search bar at the top right of the table and the as well as downloaded in a tab-delimited format:
1.3 Metadata Search
RummaGEO also provides direct metadata search of the GEO studies contained within the database. Paginated results are returned with accompanying metadata of the returned signatures: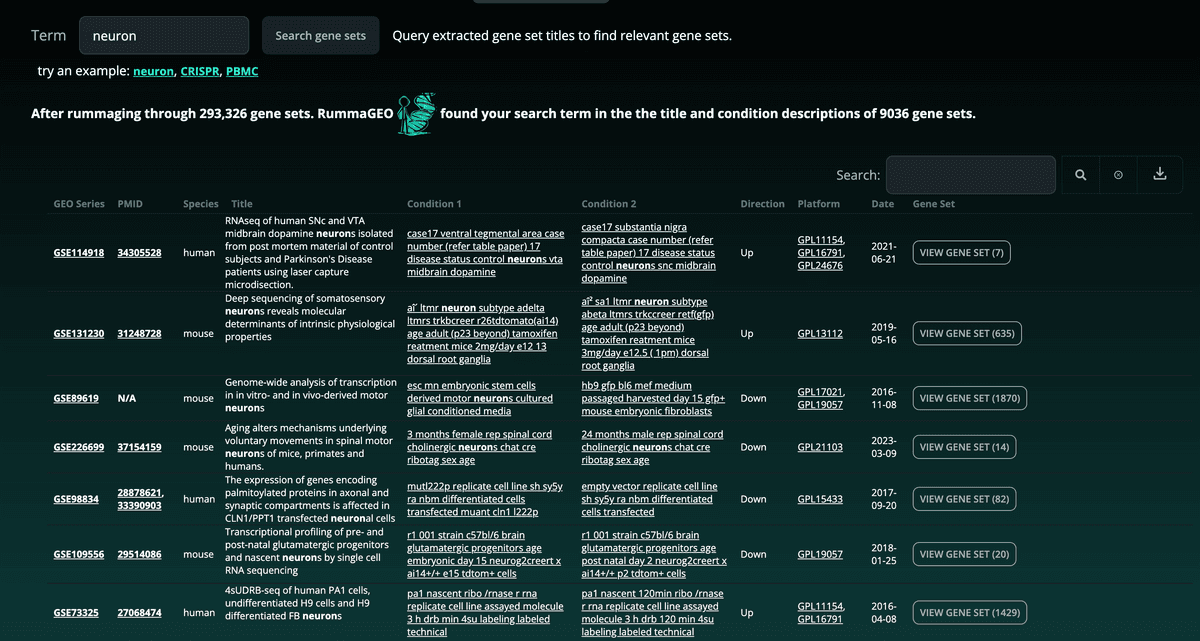 These results can also be filtered using the search bar at the top right of the table and the results table can be downloaded in a tab-delimited format:
These results can also be filtered using the search bar at the top right of the table and the results table can be downloaded in a tab-delimited format: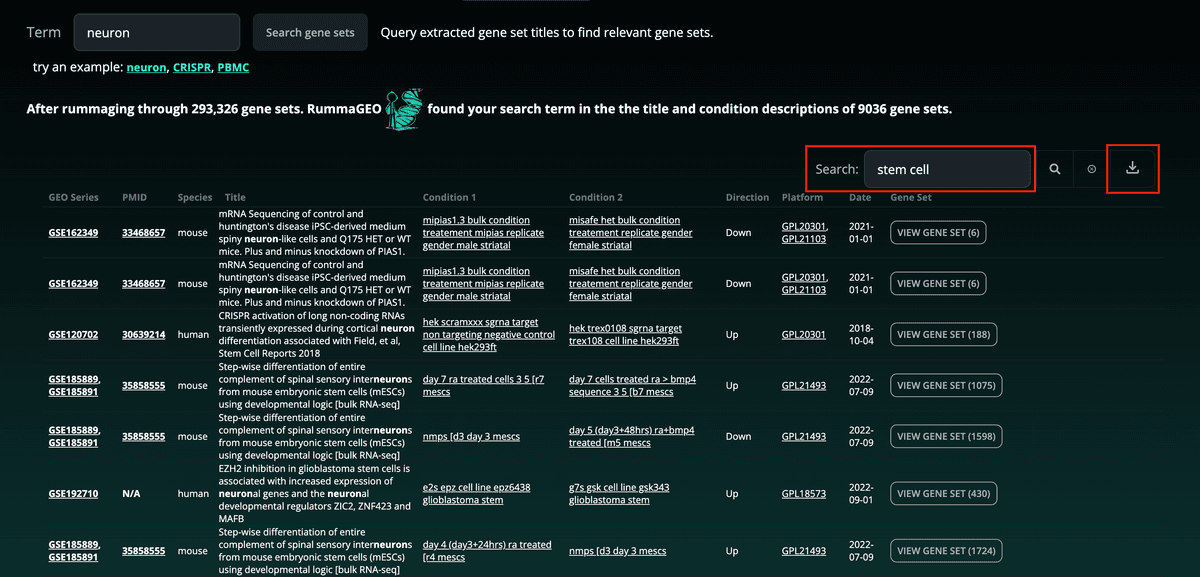
1.4 API
RummaGEO priovides programtic access through a GraphQL endpoint. Users can learn more about GraphQL queries from their provided documentation. The RummaGEO GraphQL endpoint and asscoiated Postgres database provide users with a wide range of available queries and with a user interface to test and develop these queries:
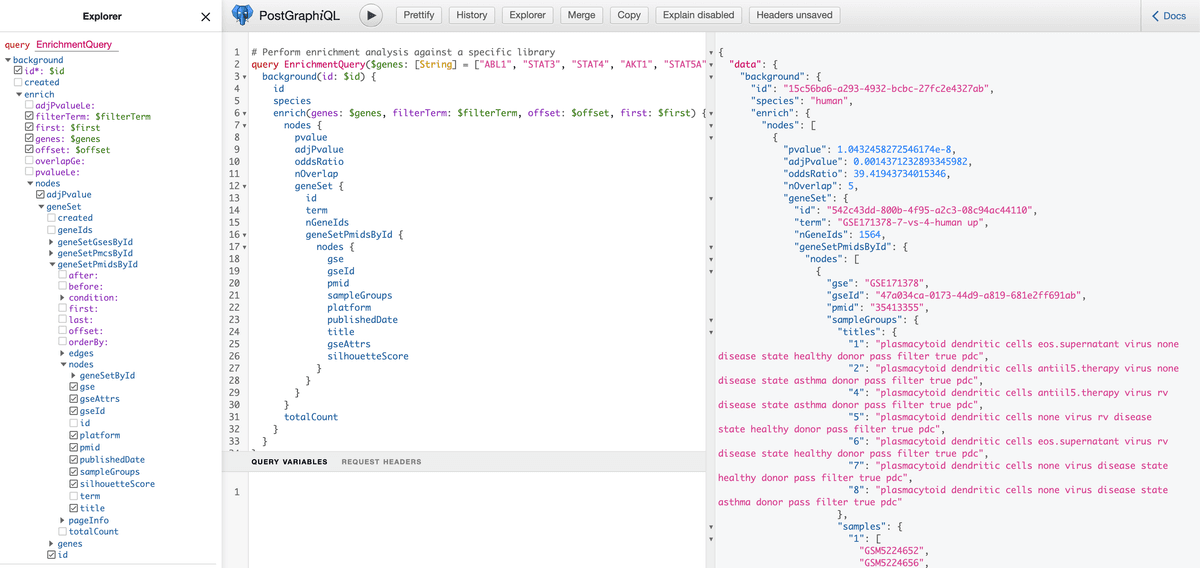
For example, enrichment analysis queries can be performed in Python agaisnt the RummaGEO human gene sets using the requests library as follows:
import requests
import json
url = "https://rummageo.com/graphql"
def enrich_rummageo(geneset: list):
query = {
"operationName": "EnrichmentQuery",
"variables": {
"filterTerm": "",
"offset": 0,
"first": 100,
"genes": geneset,
"id": "15c56ba6-a293-4932-bcbc-27fc2e4327ab"
},
"query": """query EnrichmentQuery($genes: [String]!, $filterTerm: String = "", $offset: Int = 0, $first: Int = 10, $id: UUID!) {
background(id: $id) {
id
species
enrich(genes: $genes, filterTerm: $filterTerm, offset: $offset, first: $first) {
nodes {
pvalue
adjPvalue
oddsRatio
nOverlap
geneSet {
id
term
nGeneIds
geneSetPmidsById {
nodes {
gse
gseId
pmid
sampleGroups
platform
publishedDate
title
__typename
}
__typename
}
__typename
}
__typename
}
totalCount
__typename
}
__typename
}
}
"""
}
headers = {
"Accept": "application/json",
"Content-Type": "application/json"
}
response = requests.post(url, data=json.dumps(query), headers=headers)
if response.status_code == 200:
res = response.json()
return res
This database is updated with new releases of ARCHS4.
RummaGEO is actively being developed by the Ma'ayan Lab.